paired end sequencing wikipedia
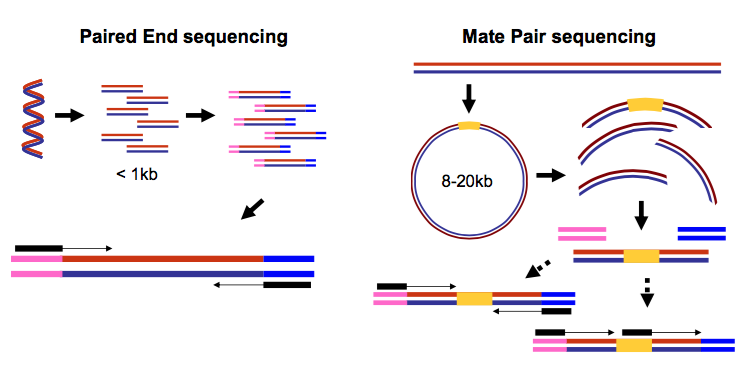
It is capable of automated paired-end reads and up to 15 Gb per run delivering over 600 bases of. Paired-end sequencing means sequencing both ends of the cDNA fragments and aligning the forward and reverse reads as read pairs Figure 8.
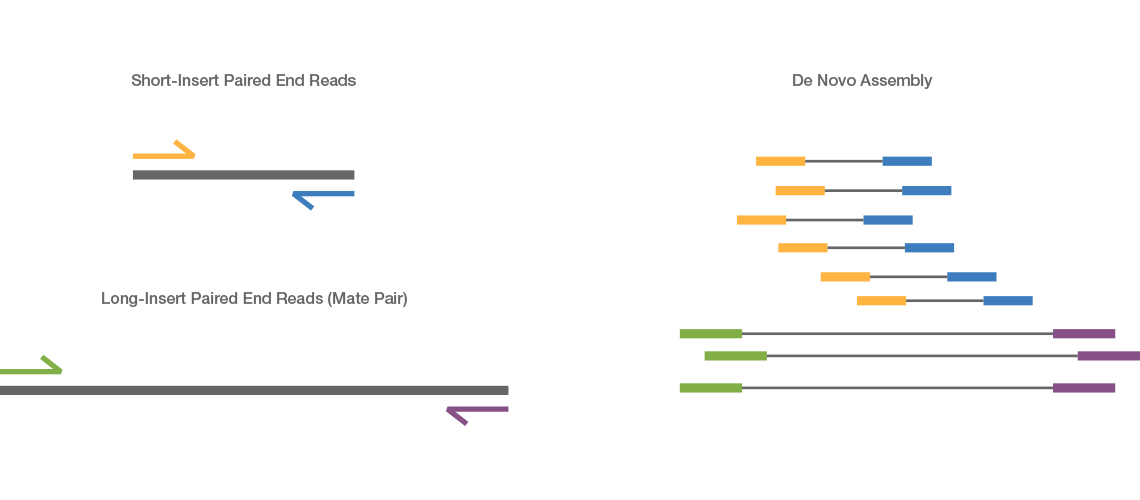
Paired-end reads are preferable for de novo.
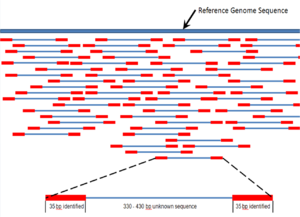
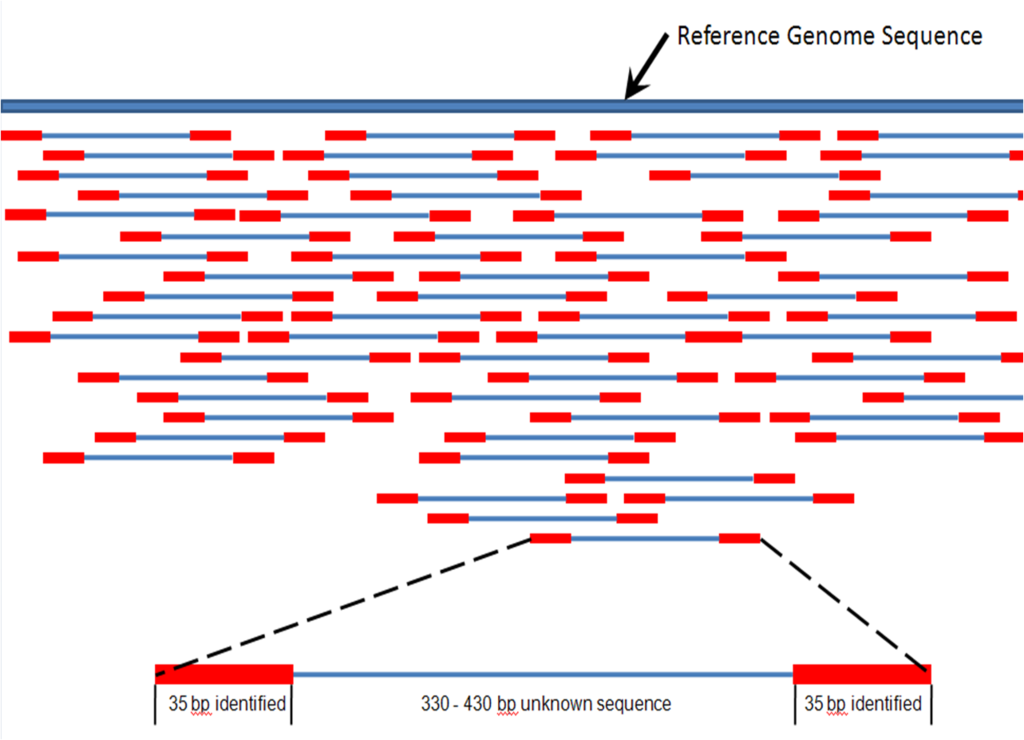
. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends. Its a subset of the first sequence starting 200 bp downstream of the first. The MiSeq System facilitates your research with a wide range of sequencing applications.
The idea is to have one of the. The second sequence is a shorter duplicate of the first sequence. Type of Run Single Read SR or Paired End PE With single read runs the sequencing instrument reads from one end of a fragment to the other end.
Paired-end sequencing facilitates detection of genomic. Paired-end or mate-pair. Paired end runs give.
Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data.

20110114 Next Generation Sequencing Course

Figure 3 From Next Generation Sequencing Platforms Semantic Scholar
File Mapping Reads Png Wikimedia Commons
Next Generation Sequencing An Overview Of The History Tools And Omic Applications Intechopen

Illumina Sequencing Technology Youtube
Manipulating Ngs Data With Galaxy Galaxy Community Hub
Rna Seq Wiki 2 Wikipedia Republished Viral Bioinformatics Research Centre

What Is Mate Pair Sequencing For

The Complete Sequence Of A Human Genome Science

Wikijournal Of Science A Broad Introduction To Rna Seq Wikiversity
Multi Omics Approaches For Comprehensive Analysis And Understanding Of The Immune Response In The Miniature Pig Breed Plos One
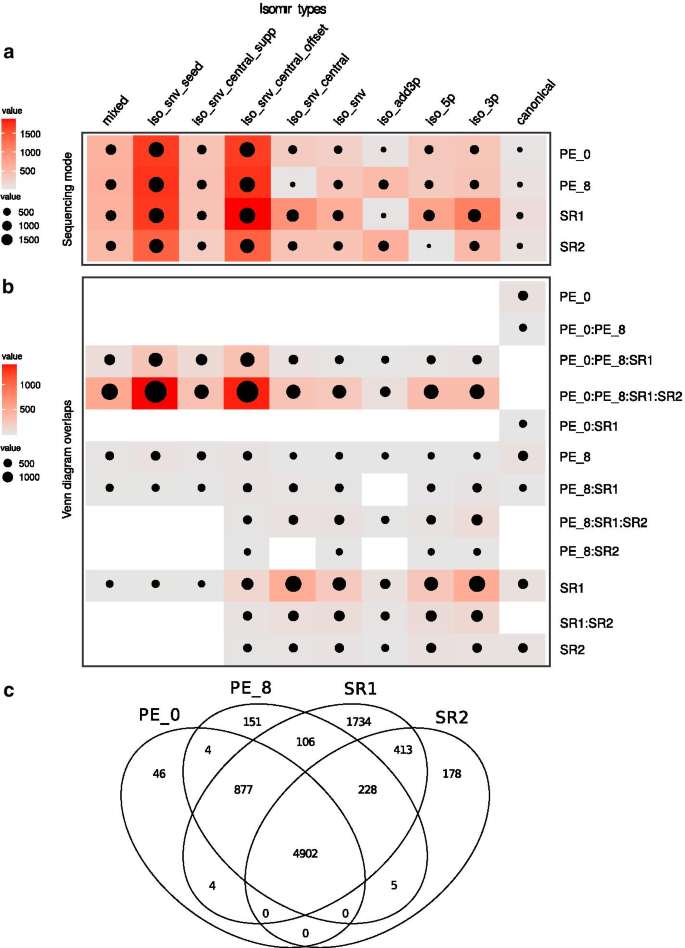
Paired End Small Rna Sequencing Reveals A Possible Overestimation In The Isomir Sequence Repertoire Previously Reported From Conventional Single Read Data Analysis Bmc Bioinformatics Full Text